Over 280 genes are involved in IRDs and can lead to isolated or syndromic disorders, i.e., associated with symptoms other than visual impairment. Similarly, over 200 genes are implicated in isolated or syndromic forms of deafness. Since the year 2000, our group has been performing molecular diagnosis of neurosensory blindness and deafness. The most common syndromic form involving an ocular-auditory phenotype is Usher syndrome (approximately 3 births per 100,000), in which congenital deafness is associated with retinitis pigmentosa (RP) that appears between early childhood and adulthood depending on the genes involved and the molecular alterations (types of pathogenic variants). Second-generation sequencing approaches and more recent third-generation techniques represent a true revolution for the molecular diagnosis of IRDs and deafness. Our group routinely sequences approximately 150 deafness and blindness genes using gene panels. Additionally, thanks to the France Genomic Medicine 2025 plan, patients with IRDs or deafness can benefit from whole-genome sequencing (WGS), with or without prior gene panel sequencing. The identification of pathogenic variants in one of the known genes, within the framework of WGS or gene panels, allows us to provide a diagnosis to the family.
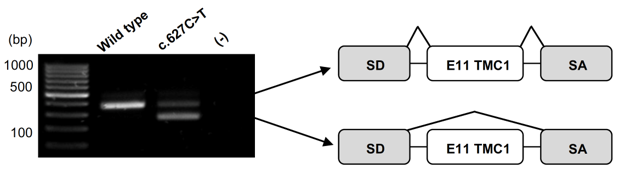
However, many variants are considered to have unknown clinical significance after analysis and interpretation with current tools, which does not allow us to provide a conclusive diagnosis to the patient. For some such variants, it is possible to assess their deleterious effect using functional analyses (especially for variants likely to alter splicing). Furthermore, it is now apparent that depending on the identified variants, some of the genes known to be involved in syndromic forms may also result in isolated forms. It is therefore necessary to develop tools to understand these different degrees of pathogenicity and to adapt patient management accordingly. As an example, genes such as USH2A and MYO7A, both implicated in Usher syndrome, have variants that lead to isolated forms, i.e., RP for USH2A and deafness for MYO7A.
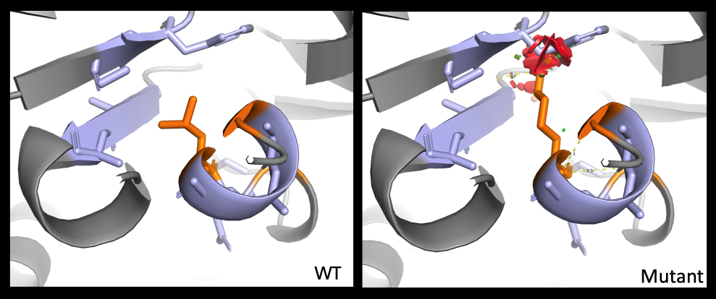
Our group focuses on the detailed characterization (bioinformatics, so-called post-genomic functional tests) of variants identified in the diagnosis of neurosensory pathologies in order to offer the patient the most comprehensive diagnosis possible, paving the way for therapeutic approaches developed within the team.
Recent publications
Vaché et al., Eur J Hum Genet, 31: 834-840, 2023
de Sainte Agathe et al., Hum Genomics, 17: 7, 2023
Mansard L et al., Diagnostics, 12: 207, 2022
Roux AF, Eur J Hum Genet, 30: 5-6, 2022
Vaché C et al., Eur J Hum Genet, 30: 34-41, 2022
Cenni C et al., Diagnostics, 11: 1636, 2022
Mansard et al., Int J Mol Sci, 22: 1324, 2021
Baux et al., Eur J Hum Genet, 29: 356-360, 2021
Fundings